UC Davis is home to faculty researchers who help advance both basic and applied knowledge about the building blocks of life.
MCBGAP scholars will be matched with faculty who share their specific interests in the biological and biomedical sciences. Potential research areas include the interplay between genetics and the environment; the dynamics, evolution, and function of the cytoskeleton; immunology of HIV; development and gene regulation; applications of imaging to studying disease; bioinformatics and genomics…and more!
Students who participate in the MCBGAP program will begin by preparing for research during the academic year via monthly videoconferences (via freely available platforms such as skype or google hangout) with their designated faculty mentors. Mentors and students will read and discuss the appropriate literature, consider possible areas of focus for summer research, and connect research preparation to the student’s coursework.
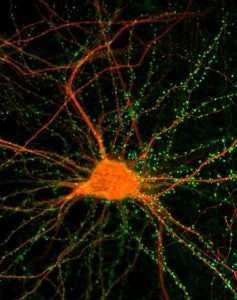

Sean Burgess explores the dynamic chromosome events that occur during the process of meiosis and how these processes are integrated to achieve accurate chromosome segregation. Chromosome missegregation is one of the leading causes of birth defects in humans. The Burgess lab combines the use of a wide array of tools, including genetics, molecular biology, biochemistry and live-cell imaging using budding yeast Saccharomyces cerevisiae and zebrafish Danio rerio as model systems.

Fred Chedin studies the molecular mechanisms of epigenetic patterning in the human genome and how these mechanisms go awry in human auto-immune disorders and cancer. Researchers in the Chedin lab aim to understand how DNA methylation patterns are established and maintained, with a focus on the role of R-loop structure formation in epigenomic patterning. Their work spans biochemical, molecular genetics, and cell culture approaches, with high-throughput genomics backed by computational analyses.

Sean Collins studies how individual cells process information, make decisions, and enact appropriate responses. He uses human neutrophils as a model system and examines how they process dynamic extracellular chemical cues to guide their behavior. Methods in his lab include a combination of techniques with particular emphasis on systematic quantitative analysis of genetic perturbations, direct monitoring of signaling using live-cell imaging of fluorescent biosensors, and mathematical modeling.

Scott Dawson examines the cytoskeleton of the intestinal parasite Giardia intestinalis. Its complex microtubule cytoskeleton is critical for motility, attachment, intracellular transport, cell division, and encystation/excystation. Rather than any specific virulence factor, the cytoskeleton itself can be considered the etiologic agent of giardiasis. Dawson’s lab also is interested in the evolution of early eukaryotes.

Elva Diaz works to understand molecular mechanisms of nervous system development in rodent model systems. Diaz and members of her lab use DNA microarrays as a tool to identify genes that are developmentally regulated during mouse brain development, and characterize candidate genes with molecular and cellular techniques and transgenic mice.

Bruce Draper studies mechanisms that regulate the development and function of germline stem cell in zebrafish, with a main focus on female germline stem cells. The Draper laboratory studies factors that function within the stem cells, as well as those that are required for the development of the somatic gonad, the germ cell niche. This, and other work, recently was profiled in a College of Biological Sciences blog post.

Joanne Engebrecht examines the role of DNA repair and checkpoint pathways in germline biology using C. elegans as a model system.An understanding of these processes in the genetically tractable worm system may provide insight into why human meiosis has a high frequency of errors resulting in human disease such as Down, Turner and Klinefelter’s Syndrome.

David Furlow‘s laboratory studies the molecular basis of hormone action, particularly during development, analyzes gene expression cascades during morphogenesis, and examines mechanisms of skeletal muscle atrophy and death.

Aldrin Gomes studies the molecular mechanisms of signal transduction, particularly in muscle contraction and cardiovascular disease. The two main research areas in the Gomes laboratory are: the role of the proteasome in normal, protected and diseased cardiac and skeletal muscles and the role of troponin in calcium regulation of muscle contraction in hypertrophic, dilated and restrictive cardiomyopathies.

John Harada uses molecular, genetic, biochemical, and genomic methods to study embryogenesis and seed development in plants.

Celina Juliano studies the molecular mechanisms that allow the freshwater cnidarian Hydra vulgaris to have “immortal” stem cells. Her research can provide insight into regenerative biology and aging in vertebrates and other organisms.

Ian Korf seeks to build better models of eukaryotic genes by investigating the individual components that define gene structure. His research uses a combination of computational modeling, comparative genomics, and experimental molecular biology to test algorithms.

Janine LaSalle examines the role of epigenetics — the study of heritable changes in chromosomes that are not encoded in the DNA sequence, including DNA methylation and chromatin organization — in human autism-spectrum disorders. Clinical applications include understanding the pathogenesis of the neurodevelopmental disorders autism, Rett syndrome, Prader-Willi syndrome, and Angelman syndrome.
See Professor LaSalle describe her work in a youtube video.

Jamal Lewis develops biomaterial systems to harness and manipulate the innate functionality of phagocytic immune cells (particularly dendritic cells) in a precise spatial and temporal manner, ultimately for therapeutic applications in immune-related conditions. These include type 1 diabetes, rheumatoid arthritis and transplant rejection.

Angelique Louie focuses on the application of engineering and physical sciences imaging principles to improve the diagnosis and management of human disease. The unifying theme of her research is the application of imaging techniques and the design of contrast agents to characterize molecular phenomena in diseased versus normal states.

Richard McKenney examins how cells internally organize using molecular motor proteins. In particular, he studies the microtubule cytoskeleton and the motor proteins that use this filament system for transport, with applications to human diseases such as cancer and neurodegeneration.
.

David Segal studies genome engineering and targeted gene regulation for applications in neurologic disorders.

Mitch Singer focuses on understanding how the bacterium Myxococcus xanthus senses nutrient limitation, and how this event initiates its developmental program.

Daniel Starr‘s lab studies processes involved in the positioning of nuclei and other organelles to specific locations within a cell. They use the nematode Caenorhabditis elegans as a model organism, with genetic, biochemical, cellular, and molecular approaches to study this basic problem in cell biology and human disease.

James Trimmer uses proteomic analysis to study cell signaling in mammalian neurons, neuronal cell biology, and membrane protein trafficking. He also directs the NIH-funded training program in molecular and cell biology.
